Xarray 101#
Sources:
Overview: Why Xarray?#
Multi-dimensional (a.k.a. N-dimensional, ND) arrays (sometimes called “tensors”) are an essential part of computational science. They are encountered in a wide range of fields, including physics, astronomy, geoscience, bioinformatics, engineering, finance, and deep learning. In Python, NumPy provides the fundamental data structure and API for working with raw ND arrays. However, real-world datasets are usually more than just raw numbers; they have labels which encode information about how the array values map to locations in space, time, etc.
Here is an example of how we might structure a dataset for a weather forecast:
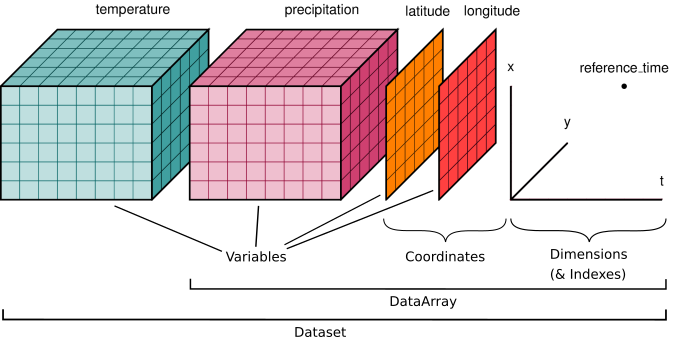
You’ll notice multiple data variables (temperature, precipitation), coordinate variables (latitude, longitude), and dimensions (x, y, t). We’ll cover how these fit into Xarray’s data structures below.
Xarray doesn’t just keep track of labels on arrays – it uses them to provide a powerful and concise interface. For example:
Apply operations over dimensions by name:
x.sum('time').Select values by label (or logical location) instead of integer location:
x.loc['2014-01-01']orx.sel(time='2014-01-01').Mathematical operations (e.g.,
x - y) vectorize across multiple dimensions (array broadcasting) based on dimension names, not shape.Easily use the split-apply-combine paradigm with groupby:
x.groupby('time.dayofyear').mean().Database-like alignment based on coordinate labels that smoothly handles missing values:
x, y = xr.align(x, y, join='outer').Keep track of arbitrary metadata in the form of a Python dictionary:
x.attrs.
The N-dimensional nature of xarray’s data structures makes it suitable for
dealing with multi-dimensional scientific data, and its use of dimension names
instead of axis labels (dim='time' instead of axis=0) makes such arrays much
more manageable than the raw numpy ndarray: with xarray, you don’t need to keep
track of the order of an array’s dimensions or insert dummy dimensions of size 1
to align arrays (e.g., using np.newaxis).
The immediate payoff of using xarray is that you’ll write less code. The long-term payoff is that you’ll understand what you were thinking when you come back to look at it weeks or months later.
Xarray’s Data structures#
Xarray provides two data structures: the DataArray and Dataset. The
DataArray class attaches dimension names, coordinates and attributes to
multi-dimensional arrays while Dataset combines multiple arrays.
Both classes are most commonly created by reading data. To learn how to create a DataArray or Dataset manually, see the Working with labeled data tutorial.
Xarray has a few small real-world tutorial datasets hosted in the pydata/xarray Github repository.
We’ll use the xarray.tutorial.load_dataset convenience function to download and open the air_temperature (National Centers for Environmental Prediction) Dataset by name.
import numpy as np
import xarray as xr
print("Numpy version:", np.__version__)
print("Xarray version:", xr.__version__)
Numpy version: 1.25.2
Xarray version: 2023.8.0
Dataset#
Dataset objects are dictionary-like containers of DataArrays, mapping a variable name to each DataArray.
ds = xr.tutorial.load_dataset("air_temperature")
ds
<xarray.Dataset>
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 241.2 242.5 243.5 ... 296.5 296.2 295.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...We can access “layers” of the Dataset (individual DataArrays) with dictionary syntax
ds["air"]
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]We can save some typing by using the “attribute” or “dot” notation. This won’t work for variable names that clash with built-in method names (for example, mean).
ds.air
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]What is all this anyway? (String representations)#
Xarray has two representation types: "html" (which is only available in
notebooks) and "text". To choose between them, use the display_style option.
So far, our notebook has automatically displayed the "html" representation (which we will continue using).
The "html" representation is interactive, allowing you to collapse sections (left arrows) and
view attributes and values for each value (right hand sheet icon and data symbol).
ds
<xarray.Dataset>
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 241.2 242.5 243.5 ... 296.5 296.2 295.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...with xr.set_options(display_style="html"):
display(ds)
<xarray.Dataset>
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 241.2 242.5 243.5 ... 296.5 296.2 295.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...The output consists of:
a summary of all Dimensions of the
Dataset(lat: 25, time: 2920, lon: 53): this tells us that the first dimension is namedlatand has a size of25, the second dimension is namedtimeand has a size of2920, and the third dimension is namedlonand has a size of53. Because we will access the dimensions by name, the order doesn’t matter.an unordered list of Coordinates or dimensions with coordinates with one item per line. Each item has a name, one or more dimensions in parentheses, a dtype and a preview of the values. Also, if it is a dimension coordinate, it will be marked with a
*or bolded like the example above.an alphabetically sorted list of Data variables, which are the actual data within the dataset. Similar to Coordinates, each data variable has a name, one or more dimensions in parentheses, a dtype and a preview of the values.
an unordered list of Indexes, which lists the indexes associated with coordinates. This is a very new feature that is part of the flexible indexes work underway. Each item has a name and a class type of index used. This currently only shows up in the “html” representation as seen above.
an unordered list of Attributes, or metadata
with xr.set_options(display_style="text"):
display(ds)
<xarray.Dataset>
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 241.2 242.5 243.5 ... 296.5 296.2 295.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...To understand each of the components better, we’ll explore the “air” variable of our Dataset.
DataArray#
The DataArray class consists of an array (data) and its associated dimension names, labels, and attributes (metadata).
da = ds["air"]
da
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]String representations#
We can use the same two representations ("html", which is only available in
notebooks, and "text") to display our DataArray.
with xr.set_options(display_style="html"):
display(da)
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]with xr.set_options(display_style="text"):
display(da)
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]In the string representation of a DataArray (versus a Dataset), we also see:
the
DataArrayname (‘air’)a preview of the array data (collapsible in the
"html"representation)
We can also access the data array directly:
ds.air.data # (or equivalently, `da.data`)
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...,
[296.4 , 295.9 , 296.19998, ..., 295.4 , 295.1 ,
294.79 ],
[296.19998, 296.69998, 296.79 , ..., 295.6 , 295.5 ,
295.1 ],
[296.29 , 297.19998, 297.4 , ..., 296.4 , 296.4 ,
296.6 ]],
[[242.29999, 242.2 , 242.29999, ..., 234.29999, 236.09999,
238.7 ],
[244.59999, 244.39 , 244. , ..., 230.29999, 232. ,
235.7 ],
[256.19998, 255.5 , 254.2 , ..., 231.2 , 233.2 ,
238.2 ],
...,
[295.6 , 295.4 , 295.4 , ..., 296.29 , 295.29 ,
295. ],
[296.19998, 296.5 , 296.29 , ..., 296.4 , 296. ,
295.6 ],
[296.4 , 296.29 , 296.4 , ..., 297. , 297. ,
296.79 ]],
...,
[[243.48999, 242.98999, 242.09 , ..., 244.18999, 244.48999,
244.89 ],
[249.09 , 248.98999, 248.59 , ..., 240.59 , 241.29 ,
242.68999],
[262.69 , 262.19 , 261.69 , ..., 239.39 , 241.68999,
245.18999],
...,
[294.79 , 295.29 , 297.49 , ..., 295.49 , 295.38998,
294.69 ],
[296.79 , 297.88998, 298.29 , ..., 295.49 , 295.49 ,
294.79 ],
[298.19 , 299.19 , 298.79 , ..., 296.09 , 295.79 ,
295.79 ]],
[[245.79 , 244.79 , 243.48999, ..., 243.29 , 243.98999,
244.79 ],
[249.89 , 249.29 , 248.48999, ..., 241.29 , 242.48999,
244.29 ],
[262.38998, 261.79 , 261.29 , ..., 240.48999, 243.09 ,
246.89 ],
...,
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Named dimensions#
.dims are the named axes of your data. They may (dimension coordinates) or may not (dimensions without coordinates) have associated values. Names can be anything that fits into a Python set (i.e. calling hash() on it doesn’t raise an error), but to be
useful they should be strings.
In this case we have 2 spatial dimensions (latitude and longitude are stored with shorthand names lat and lon) and one temporal dimension (time).
ds.air.dims
('time', 'lat', 'lon')
Coordinates#
.coords is a simple dict-like data container
for mapping coordinate names to values. These values can be:
another
DataArrayobjecta tuple of the form
(dims, data, attrs)whereattrsis optional. This is roughly equivalent to creating a newDataArrayobject withDataArray(dims=dims, data=data, attrs=attrs)a 1-dimensional
numpyarray (or anything that can be coerced to one usingnumpy.array, such as alist) containing numbers, datetime objects, strings, etc. to label each point.
Here we see the actual timestamps and spatial positions of our air temperature data:
ds.air.coords
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Note
The difference between the dimension labels (dimension coordinates) and normal
coordinates is that for now it only is possible to use indexing operations
(sel, reindex, etc.) with dimension coordinates. Also, while coordinates can
have arbitrary dimensions, dimension coordinates have to be one-dimensional.
Attributes#
.attrs is a dictionary that can contain arbitrary Python objects (strings, lists, integers, dictionaries, etc.) containing information about your data. Your only
limitation is that some attributes may not be writeable to certain file formats.
ds.air.attrs
{'long_name': '4xDaily Air temperature at sigma level 995',
'units': 'degK',
'precision': 2,
'GRIB_id': 11,
'GRIB_name': 'TMP',
'var_desc': 'Air temperature',
'dataset': 'NMC Reanalysis',
'level_desc': 'Surface',
'statistic': 'Individual Obs',
'parent_stat': 'Other',
'actual_range': array([185.16, 322.1 ], dtype=float32)}
To Pandas and back#
DataArray and Dataset objects are frequently created by converting from
other libraries such as pandas or by reading from
data storage formats such as
NetCDF or
zarr.
To convert from / to pandas, we can use the
to_xarray
methods on pandas objects or the
to_pandas
methods on xarray objects:
import pandas as pd
print("Pandas version:", pd.__version__)
Pandas version: 2.0.3
series = pd.Series(np.ones((10,)), index=list("abcdefghij"))
series
a 1.0
b 1.0
c 1.0
d 1.0
e 1.0
f 1.0
g 1.0
h 1.0
i 1.0
j 1.0
dtype: float64
arr = series.to_xarray()
arr
<xarray.DataArray (index: 10)> array([1., 1., 1., 1., 1., 1., 1., 1., 1., 1.]) Coordinates: * index (index) object 'a' 'b' 'c' 'd' 'e' 'f' 'g' 'h' 'i' 'j'
arr.to_pandas()
index
a 1.0
b 1.0
c 1.0
d 1.0
e 1.0
f 1.0
g 1.0
h 1.0
i 1.0
j 1.0
dtype: float64
We can also control what pandas object is used by calling to_series or
to_dataframe:
to_series will always convert DataArray objects to
pandas.Series, using a MultiIndex for higher dimensions
ds.air.to_series()
time lat lon
2013-01-01 00:00:00 75.0 200.0 241.199997
202.5 242.500000
205.0 243.500000
207.5 244.000000
210.0 244.099991
...
2014-12-31 18:00:00 15.0 320.0 297.389984
322.5 297.190002
325.0 296.489990
327.5 296.190002
330.0 295.690002
Name: air, Length: 3869000, dtype: float32
to_dataframe will always convert DataArray or Dataset
objects to a pandas.DataFrame. Note that DataArray objects have to be named
for this.
ds.air.to_dataframe()
| air | |||
|---|---|---|---|
| time | lat | lon | |
| 2013-01-01 00:00:00 | 75.0 | 200.0 | 241.199997 |
| 202.5 | 242.500000 | ||
| 205.0 | 243.500000 | ||
| 207.5 | 244.000000 | ||
| 210.0 | 244.099991 | ||
| ... | ... | ... | ... |
| 2014-12-31 18:00:00 | 15.0 | 320.0 | 297.389984 |
| 322.5 | 297.190002 | ||
| 325.0 | 296.489990 | ||
| 327.5 | 296.190002 | ||
| 330.0 | 295.690002 |
3869000 rows × 1 columns
Since columns in a DataFrame need to have the same index, they are broadcasted.
ds.to_dataframe()
| air | |||
|---|---|---|---|
| lat | time | lon | |
| 75.0 | 2013-01-01 00:00:00 | 200.0 | 241.199997 |
| 202.5 | 242.500000 | ||
| 205.0 | 243.500000 | ||
| 207.5 | 244.000000 | ||
| 210.0 | 244.099991 | ||
| ... | ... | ... | ... |
| 15.0 | 2014-12-31 18:00:00 | 320.0 | 297.389984 |
| 322.5 | 297.190002 | ||
| 325.0 | 296.489990 | ||
| 327.5 | 296.190002 | ||
| 330.0 | 295.690002 |
3869000 rows × 1 columns
Indexing and Selecting Data#
Xarray offers extremely flexible indexing routines that combine the best features of NumPy and Pandas for data selection.
The most basic way to access elements of a DataArray object is to use Python’s [] syntax, such as array[i, j], where i and j are both integers.
As xarray objects can store coordinates corresponding to each dimension of an array, label-based indexing is also possible (e.g. .sel(latitude=0), similar to pandas.DataFrame.loc). In label-based indexing, the element position i is automatically looked-up from the coordinate values.
By leveraging the labeled dimensions and coordinates provided by Xarray, users can effortlessly access, subset, and manipulate data along multiple axes, enabling complex operations such as slicing, masking, and aggregating data based on specific criteria.
This indexing and selection capability of Xarray not only enhances data exploration and analysis workflows but also promotes reproducibility and efficiency by providing a convenient interface for working with multi-dimensional data structures.
In total, xarray supports four different kinds of indexing, as described below and summarized in this table:
Dimension lookup |
Index lookup |
|
|
|---|---|---|---|
Positional |
By integer |
|
not available |
Positional |
By label |
|
not available |
By name |
By integer |
|
|
By name |
By label |
|
|
Label-based indexing#
Select data by coordinate labels. Xarray inherits its label-based indexing rules from pandas; this means great support for dates and times!
# pull out data for all of 2013-May
ds.sel(time="2013-05")
<xarray.Dataset>
Dimensions: (lat: 25, time: 124, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-05-01 ... 2013-05-31T18:00:00
Data variables:
air (time, lat, lon) float32 259.2 259.3 259.1 ... 298.2 297.6 297.5
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...# slicing data from May to July 2013
ds.sel(time=slice("2013-05", "2013-07"))
<xarray.Dataset>
Dimensions: (lat: 25, time: 368, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-05-01 ... 2013-07-31T18:00:00
Data variables:
air (time, lat, lon) float32 259.2 259.3 259.1 ... 299.4 299.5 299.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...# can also get the whole year
ds.sel(time="2013")
<xarray.Dataset>
Dimensions: (lat: 25, time: 1460, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2013-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 241.2 242.5 243.5 ... 296.1 295.1 294.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...# demonstrate "nearest" indexing
ds.sel(lon=240.2, method="nearest")
<xarray.Dataset>
Dimensions: (lat: 25, time: 2920)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
lon float32 240.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat) float32 239.6 237.2 240.1 249.0 ... 294.8 296.9 298.4
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...# "nearest indexing at multiple points"
ds.sel(lon=[240.125, 234], lat=[40.3, 50.3], method="nearest")
<xarray.Dataset>
Dimensions: (lat: 2, time: 2920, lon: 2)
Coordinates:
* lat (lat) float32 40.0 50.0
* lon (lon) float32 240.0 235.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 268.1 283.0 265.5 ... 285.2 256.8 268.6
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...Position-based indexing#
This is similar to your usual numpy array[0, 2, 3] but with the power of named dimensions!
ds.air.data[0, 2, 3]
247.5
# pull out time index 0, lat index 2, and lon index 3
ds.air.isel(time=0, lat=2, lon=3) # much better than ds.air[0, 2, 3]
<xarray.DataArray 'air' ()>
array(247.5, dtype=float32)
Coordinates:
lat float32 70.0
lon float32 207.5
time datetime64[ns] 2013-01-01
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]# demonstrate slicing
ds.air.isel(lat=slice(10))
<xarray.DataArray 'air' (time: 2920, lat: 10, lon: 53)>
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[274.79 , 275.19998, 275.6 , ..., 277.19998, 277. ,
277. ],
[275.9 , 276.9 , 276.9 , ..., 280.9 , 280.5 ,
279.69998],
[276.69998, 277.4 , 277.69998, ..., 283.29 , 284.1 ,
283.9 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...
[275.59 , 276.29 , 277.49 , ..., 275.19 , 275.79 ,
276.59 ],
[276.88998, 277.88998, 278.69 , ..., 273.59 , 274.29 ,
275.29 ],
[276.79 , 277.29 , 278.29 , ..., 274.19 , 275.38998,
276.88998]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[274.29 , 274.49 , 275.59 , ..., 274.69 , 274.99 ,
275.38998],
[276.79 , 277.49 , 277.99 , ..., 273.19 , 273.59 ,
274.19 ],
[276.88998, 277.29 , 277.59 , ..., 273.79 , 274.99 ,
276.19 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 62.5 60.0 57.5 55.0 52.5
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]Concepts for computation#
Here is a motivating calculation where we subtract two DataArrays with data available at different locations in the (space, time) plane.
arr1 = xr.DataArray(
np.arange(12).reshape(3, 4),
dims=("space", "time"),
coords={"space": ["a", "b", "c"], "time": [0, 1, 2, 3]},
)
arr1
<xarray.DataArray (space: 3, time: 4)>
array([[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]])
Coordinates:
* space (space) <U1 'a' 'b' 'c'
* time (time) int64 0 1 2 3arr2 = xr.DataArray(
[0, 1],
dims="space",
coords={"space": ["b", "d"]},
)
arr2
<xarray.DataArray (space: 2)> array([0, 1]) Coordinates: * space (space) <U1 'b' 'd'
Note
arr1 is 2D; while arr2 is 1D along space and has values at two locations only.
arr1 - arr2
<xarray.DataArray (space: 1, time: 4)> array([[4, 5, 6, 7]]) Coordinates: * space (space) <U1 'b' * time (time) int64 0 1 2 3
To understand this output, we must understand two fundamental concepts underlying computation with Xarray objects
Broadcasting: The objects need to have compatible shapes.
Alignment: The objects need to have values at the same coordinate labels
Broadcasting: adjusting arrays to the same shape#
Broadcasting allows an operator or a function to act on two or more arrays to operate even if these arrays do not have the same shape. That said, not all the dimensions can be subjected to broadcasting; they must meet certain rules. The image below illustrates how an operation on arrays with different coordinates will result in automatic broadcasting
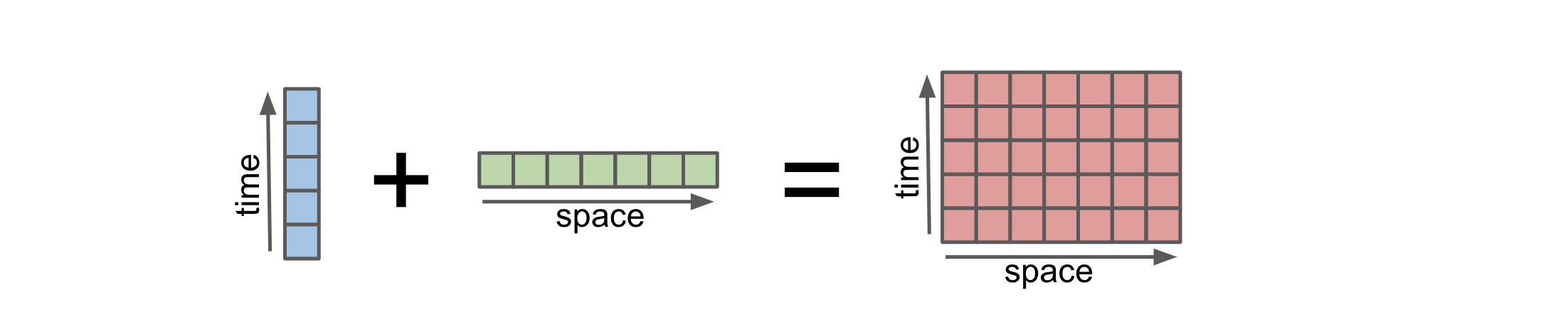
Credit: Stephan Hoyer – xarray ECMWF Python workshop
Numpy’s broadcasting rules, based on array shape, can sometimes be difficult to understand and remember. Xarray does broadcasting by dimension name, rather than array shape. This is a huge convenience.
Here are two 1D arrays
array1 = xr.DataArray(
np.arange(3),
dims="space",
coords={"space": ["a", "b", "c"]},
name="array1",
)
array2 = xr.DataArray(
np.arange(4),
dims="time",
coords={"time": [0, 1, 2, 3]},
name="array2",
)
display(array1)
display(array2)
<xarray.DataArray 'array1' (space: 3)> array([0, 1, 2]) Coordinates: * space (space) <U1 'a' 'b' 'c'
<xarray.DataArray 'array2' (time: 4)> array([0, 1, 2, 3]) Coordinates: * time (time) int64 0 1 2 3
Let’s subtract the two:
array1 - array2
<xarray.DataArray (space: 3, time: 4)>
array([[ 0, -1, -2, -3],
[ 1, 0, -1, -2],
[ 2, 1, 0, -1]])
Coordinates:
* space (space) <U1 'a' 'b' 'c'
* time (time) int64 0 1 2 3We see that the result is a 2D array.
When subtracting, Xarray first realizes that array1 is missing the dimension time and array2 is missing the dimension space. Xarray then broadcasts or “expands” both arrays to 2D with dimensions space, time. Here is an illustration:

Broadcasting in numpy#
For contrast let us examine the pure numpy version of this calculation. We use .data to extract the underlying numpy array object.
array1.data - array2.data
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[37], line 1
----> 1 array1.data - array2.data
ValueError: operands could not be broadcast together with shapes (3,) (4,)
To get this calculation to work, we need to insert new axes manually using np.newaxis.
array1.data[:, np.newaxis] - array2.data[np.newaxis, :]
array([[ 0, -1, -2, -3],
[ 1, 0, -1, -2],
[ 2, 1, 0, -1]])
Because xarray knows about dimension names we avoid having to create unnecessary
size-1 dimensions using np.newaxis or .reshape. This is yet another example where the metadata (dimension names) reduces the mental overhead associated with coding a calculation
For more, see the Xarray documentation and the numpy documentation on broadcasting.
Alignment: putting data on the same grid#
When combining two input arrays using an arithmetic operation, both arrays must first be converted to the same coordinate system. This is “alignment”.
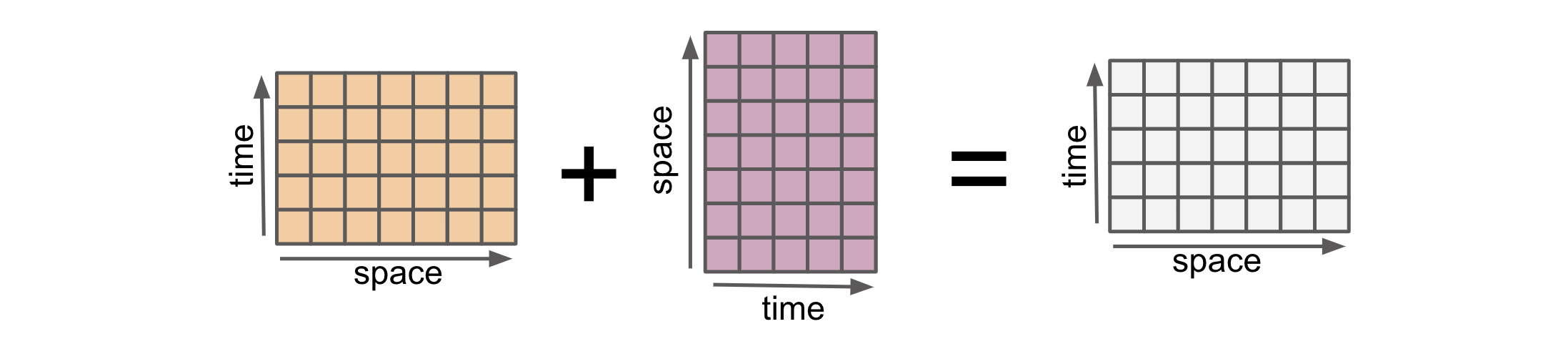
Here are two 2D DataArrays with different shapes.
arr1 = xr.DataArray(
np.arange(12).reshape(3, 4),
dims=("space", "time"),
coords={"space": ["a", "b", "c"], "time": [0, 1, 2, 3]},
)
arr1
<xarray.DataArray (space: 3, time: 4)>
array([[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]])
Coordinates:
* space (space) <U1 'a' 'b' 'c'
* time (time) int64 0 1 2 3arr2 = xr.DataArray(
np.arange(14).reshape(2, 7),
dims=("space", "time"),
coords={"space": ["b", "d"], "time": [-2, -1, 0, 1, 2, 3, 4]},
)
arr2
<xarray.DataArray (space: 2, time: 7)>
array([[ 0, 1, 2, 3, 4, 5, 6],
[ 7, 8, 9, 10, 11, 12, 13]])
Coordinates:
* space (space) <U1 'b' 'd'
* time (time) int64 -2 -1 0 1 2 3 4arr1 and arr2 have the same dimensions (space, time) but have values at different locations in the (space, time) plane with some locations in common.
Note
xarray assumes coordinate labels are in the same coordinate system such that space=’b’ in arr1 is the same as space=’b’ in arr2. For more sophisticated handling of coordinate systems see rioxarray
We see that both arrays only have values in common at x=”b” and y=[0, 1, 2, 3]. Before applying an arithmetic operation we must first modify each DataArray so that they have values at the same points. This is “alignment”.
Controlling alignment#
We can explicitly align objects using xr.align. The key decision to make is how to decide which points must be kept. The other way to think of alignment is that objects must be converted to a common grid prior to any operation combining multiiple objects. This decision is controlled by the "join" keyword argument. Xarray provides 5 ways to convert the coordinate labels of multiple Datasets to a common grid. This terminology originates in the database community.
join="inner"or reindex to the “intersection set” of coordinate labelsjoin="outer"or reindex to the “union set” of coordinate labelsjoin="left"or reindex to the coordinate labels of the leftmost objectjoin="right"or reindex to the coordinate labels of the rightmost objectjoin="exact"checks for exact equality of coordinate labels before the operation.
First lets try an inner join. This is the default for arithmetic operations in Xarray. We see that the result has values for locations that arr1 and arr2 have in common: x="b" and y=[0, 1, 2, 3]. Here is an illustration

a1_aligned, a2_aligned = xr.align(arr1, arr2, join="inner")
display(a1_aligned)
display(a2_aligned)
<xarray.DataArray (space: 1, time: 4)> array([[4, 5, 6, 7]]) Coordinates: * space (space) <U1 'b' * time (time) int64 0 1 2 3
<xarray.DataArray (space: 1, time: 4)> array([[2, 3, 4, 5]]) Coordinates: * space (space) <U1 'b' * time (time) int64 0 1 2 3
High level computation#
(groupby, resample, rolling, coarsen, weighted)
Xarray has some very useful high level objects that let you do common computations:
groupby: Bin data in to groups and reduceresample: Groupby specialized for time axes. Either downsample or upsample your data.rolling: Operate on rolling windows of your data e.g. running meancoarsen: Downsample your dataweighted: Weight your data before reducing
Below we quickly demonstrate these patterns. See the user guide links above and the tutorial for more.
groupby#
Xarray copies Pandas’ very useful groupby functionality, enabling the “split / apply / combine” workflow on xarray DataArrays and Datasets.
# seasonal groups
ds.groupby("time.season")
DatasetGroupBy, grouped over 'season'
4 groups with labels 'DJF', 'JJA', 'MAM', 'SON'.
# make a seasonal mean
seasonal_mean = ds.groupby("time.season").mean()
seasonal_mean
<xarray.Dataset>
Dimensions: (lat: 25, season: 4, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* season (season) object 'DJF' 'JJA' 'MAM' 'SON'
Data variables:
air (season, lat, lon) float32 247.0 247.0 246.7 ... 299.4 299.4 299.5
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...The seasons are out of order (they are alphabetically sorted). This is a common
annoyance. The solution is to use .sel to change the order of labels
seasonal_mean = seasonal_mean.sel(season=["DJF", "MAM", "JJA", "SON"])
seasonal_mean
<xarray.Dataset>
Dimensions: (lat: 25, season: 4, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* season (season) object 'DJF' 'MAM' 'JJA' 'SON'
Data variables:
air (season, lat, lon) float32 247.0 247.0 246.7 ... 299.4 299.4 299.5
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...resample#
Resampling means changing the time frequency of data, usually reducing to a coarser frequency: e.g. converting daily frequency data to monthly frequency data using mean to reduce the values. This operation can be thought of as a groupby operation where each group is a single month of data. Resampling can be applied only to time-index dimensions.
# resample to monthly frequency
ds.resample(time="M").mean()
<xarray.Dataset>
Dimensions: (lat: 25, time: 24, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-31 2013-02-28 ... 2014-12-31
Data variables:
air (time, lat, lon) float32 244.5 244.7 244.7 ... 297.7 297.7 297.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...weighted#
Xarray supports weighted array reductions
DataArray and Dataset objects include DataArray.weighted() and Dataset.weighted() array reduction methods. They currently support weighted sum, mean, std, var and quantile.
coords = dict(month=("month", [1, 2, 3]))
prec = xr.DataArray([1.1, 1.0, 0.9], dims=("month",), coords=coords)
weights = xr.DataArray([31, 28, 31], dims=("month",), coords=coords)
# Created weighted object
weighted_prec = prec.weighted(weights)
weighted_prec
DataArrayWeighted with weights along dimensions: month
# Calculate the weighted sum
weighted_prec.sum()
<xarray.DataArray ()> array(90.)
# Calculate the weighted mean
weighted_prec.mean(dim="month")
<xarray.DataArray ()> array(1.)
# Calculate the weighted quantile
weighted_prec.quantile(q=0.5, dim="month")
<xarray.DataArray ()>
array(1.)
Coordinates:
quantile float64 0.5Note
weights must be a DataArray and cannot contain missing values. Missing values can be replaced manually by weights.fillna(0).
Visualization#
Xarray lets you easily visualize datasets easily by default and integrates really well with the Holoviz ecosystem.
Plot the air temperature seasonal mean from above groupby
seasonal_mean.air.plot(col="season", col_wrap=2);
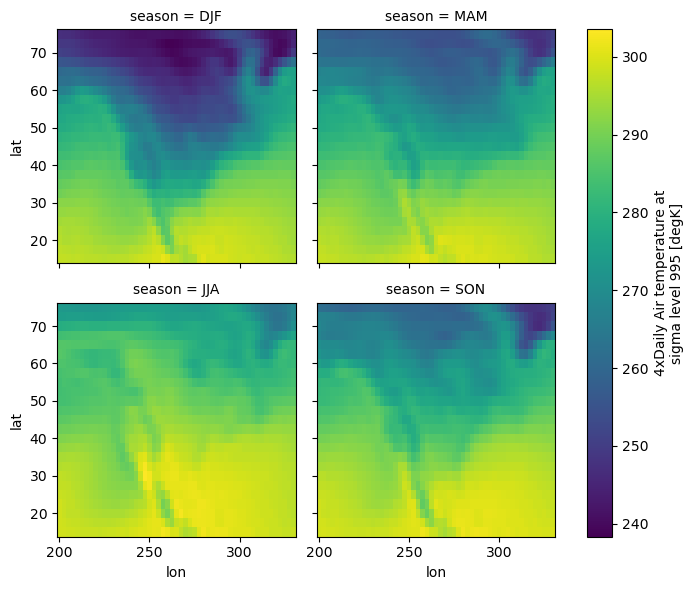
# contours
seasonal_mean.air.plot.contour(col="season", levels=20, add_colorbar=True);
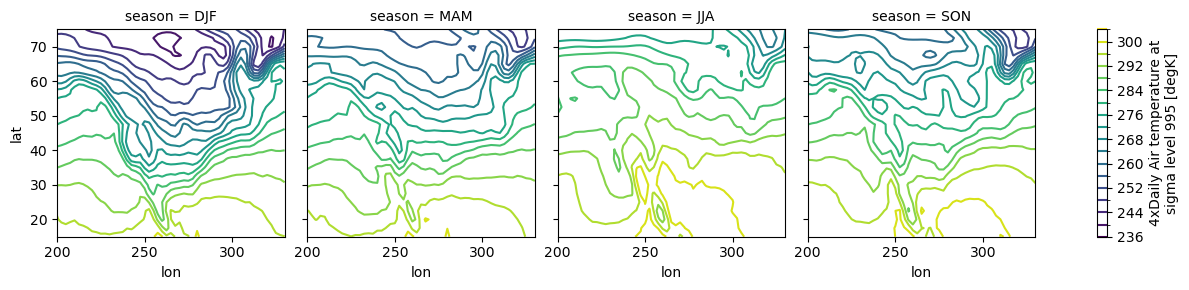
# line plots as well
seasonal_mean.air.mean("lon").plot.line(hue="season", y="lat");
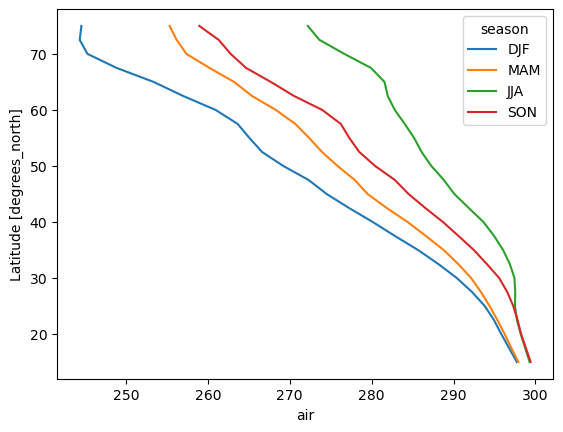
For more see the user guide, the gallery, and the tutorial material.
Reading and writing files#
Xarray supports many disk formats. Below is a small example using netCDF. For more see the documentation
ds.to_netcdf("my-example-dataset.nc")
/tmp/ipykernel_1880/2633684323.py:1: SerializationWarning: saving variable air with floating point data as an integer dtype without any _FillValue to use for NaNs
ds.to_netcdf("my-example-dataset.nc")
Note
To avoid the SerializationWarning you can assign a _FillValue for any NaNs in ‘air’ array by adding the keyword argument encoding=dict(air={_FillValue=-9999})
# read from disk
fromdisk = xr.open_dataset("my-example-dataset.nc")
fromdisk
<xarray.Dataset>
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 ...
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...# check that the two are identical
ds.identical(fromdisk)
True
Tip
A common use case to read datasets that are a collection of many netCDF files. See the documentation for how to handle that.
Finally to read other file formats, you might find yourself reading in the data using a different library and then creating a DataArray(docs, tutorial) from scratch. For example, you might use h5py to open an HDF5 file and then create a Dataset from that.
For MATLAB files you might use scipy.io.loadmat or h5py depending on the version of MATLAB file you’re opening and then construct a Dataset.
What’s Next#
Read the tutorial material and user guide
See the description of common terms used in the xarray documentation
Answers to common questions on “how to do X” with Xarray are here
Ryan Abernathey has a book on data analysis with a chapter on Xarray
Project Pythia has foundational and more advanced material on Xarray. Pythia also aggregates other Python learning resources.
The Xarray Github Discussions and Pangeo Discourse are good places to ask questions.
Tell your friends! Tweet!
